Convex gating is a Python package to infer an optimal gating strategy from flow, cyTOF or Ab/CITE-seq data. Convex gating expects a labelled input (for instance, from clustering) and returns a gating panel to separate the selected group of events (e.g. a cluster) from all other events (see Fig. 1a). For each cluster, it reports the purity (precision), yield (recall) and the harmonic mean of both metrics (F1 score) for each gate hierarchy and the entire gating strategy. It relies on the scanpy/anndata for the data format and data pre-processing and further on PyTorch for stochastic gradient descent. Therefore, resulting gates may slightly vary.
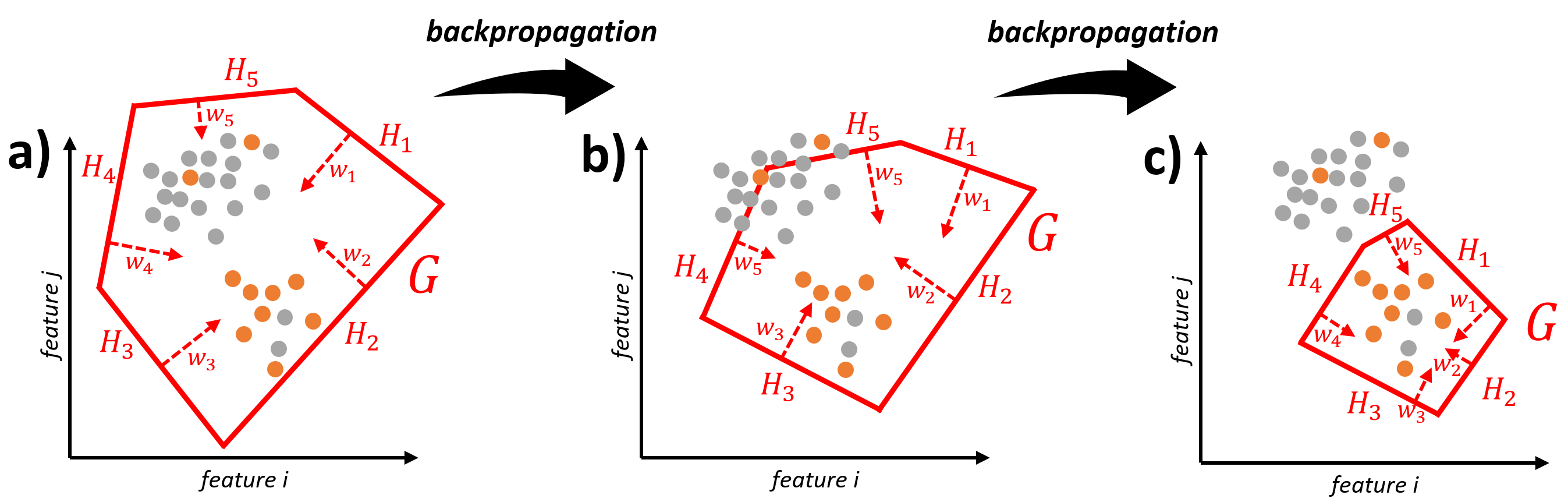
The iterative procedure to find a suitable gate before applying the convex hull is illustrated in the following graphic.
git clone https://github.com/buettnerlab/convexgating.git
cd convexgating
pip install -e .Generating gating strategy for cells or events labeled 'cluster_A' from an AnnData object (adata) using labels found in adata.obs[cluster_string].
import convexgating as cg
import scanpy as sc
adata = sc.read_h5ad(adata_path)
cluster_string = 'clustering' #column in adata.obs
target_cluster = 'cluster_A' #category in adata.obs[cluster_string]
save = save_path
cg.tools.CONVEX_GATING(adata=adata,
cluster_numbers = [target_cluster],
cluster_string = cluster_string,
save_path=save)Generating a series of gating strategies for cells or events labeled 'cluster_A','cluster_B','cluster_C' from an AnnData object (adata) using labels found in adata.obs[cluster_string].
import convexgating as cg
import scanpy as sc
adata = sc.read_h5ad(adata_path)
cluster_string = 'clustering' #column in adata.obs
target_clusters = ['cluster_A','cluster_B','cluster_C'] #categories in adata.obs[cluster_string]
save = save_path
cg.tools.CONVEX_GATING(adata=adata,
cluster_numbers = target_clusters,
cluster_string = cluster_string,
save_path=save)A usage example is available in the docs/tutorials/tutorial_01.ipynb file located in this repository.
Please see the Command-line Reference for further details.
This package was created with cookietemple using Cookiecutter based on Hypermodern_Python_Cookiecutter.